News Center
News and Development
Literature Sharing: Visual Identification and Typing of Toxigenic Vibrio cholerae O1/O139 Serogroups Using CARID Technology
Release Time:
2025-09-23
The CARID system represents a significant advancement in the rapid, sensitive, and field-deployable detection of toxigenic Vibrio cholerae.
According to data from the World Health Organization (WHO), as of 2025, cholera remains a significant public health challenge in multiple regions globally. From January 1 to August 17, 2025, a total of 31 countries reported 409,222 cases of cholera/acute watery diarrhea (AWD), including 4,738 deaths.
To enhance early case detection and cholera outbreak surveillance, WHO recommends the use of Rapid Diagnostic Tests (RDTs) for the early identification of suspected or potential cholera outbreaks.
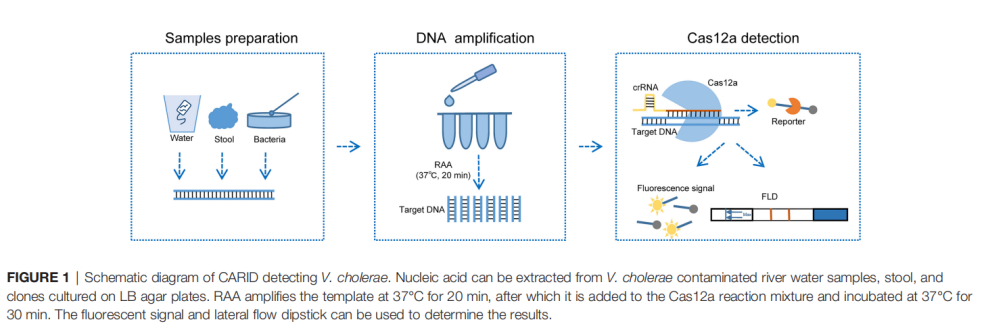
In this context, this article shares a recent study that developed a Cas12a-assisted rapid isothermal detection (CARID) system capable of detecting toxigenic Vibrio cholerae O1 and O139 serotypes within one hour.
Background
Cholera remains a major threat to global public health, particularly in areas with poor sanitation. Cholera toxin (CT) is the key virulence factor of V. cholerae, encoded by the ctxA and ctxB genes. Strains carrying these genes are defined as toxigenic. Although V. cholerae has over 200 serogroups, only the toxigenic O1 and O139 serogroups are associated with large-scale epidemics and pandemics.
Limitations of Existing Detection Methods
Traditional culture-based methods for identifying V. cholerae require at least three days and rely on advanced laboratory infrastructure and skilled personnel. While molecular techniques like PCR and qPCR are faster, they still require expensive equipment and controlled laboratory environments, making them less suitable for resource-limited settings.
Therefore, there is an urgent need for rapid, point-of-care testing methods to facilitate the timely detection and control of cholera.
Technological Innovation: The CARID System
The CARID system integrates two cutting-edge biotechnologies:
• CRISPR-Cas12a: The CRISPR-Cas12a system, guided by an in vitro crRNA, binds to the target sequence and subsequently exhibits collateral activity, indiscriminately cleaving single-stranded DNA (ssDNA). This property has been harnessed for specific pathogen detection. For double-stranded DNA (dsDNA) targets, effective cleavage of non-target DNA by Cas12a requires recognition of a short T-rich sequence (5'-TTTN-3') in the target strand, known as the protospacer adjacent motif (PAM). This recognition mechanism provides theoretical guidance for crRNA design.
• RAA Technology (Recombinase Aided Amplification): This is an isothermal amplification technique that can rapidly amplify target DNA at 37–42°C within 15–30 minutes, without the need for complex equipment like thermal cyclers.
Result Interpretation Methods
The CARID system supports two result readout methods:
• Fluorescence Detection: Cleavage of an ssDNA reporter molecule releases a fluorescent signal, measurable with a portable fluorometer.
• Lateral Flow Dipstick (LFD): Results are interpreted based on the appearance of a test line on a strip, visible to the naked eye—making it highly suitable for field applications. No instrument is needed.
Performance Evaluation
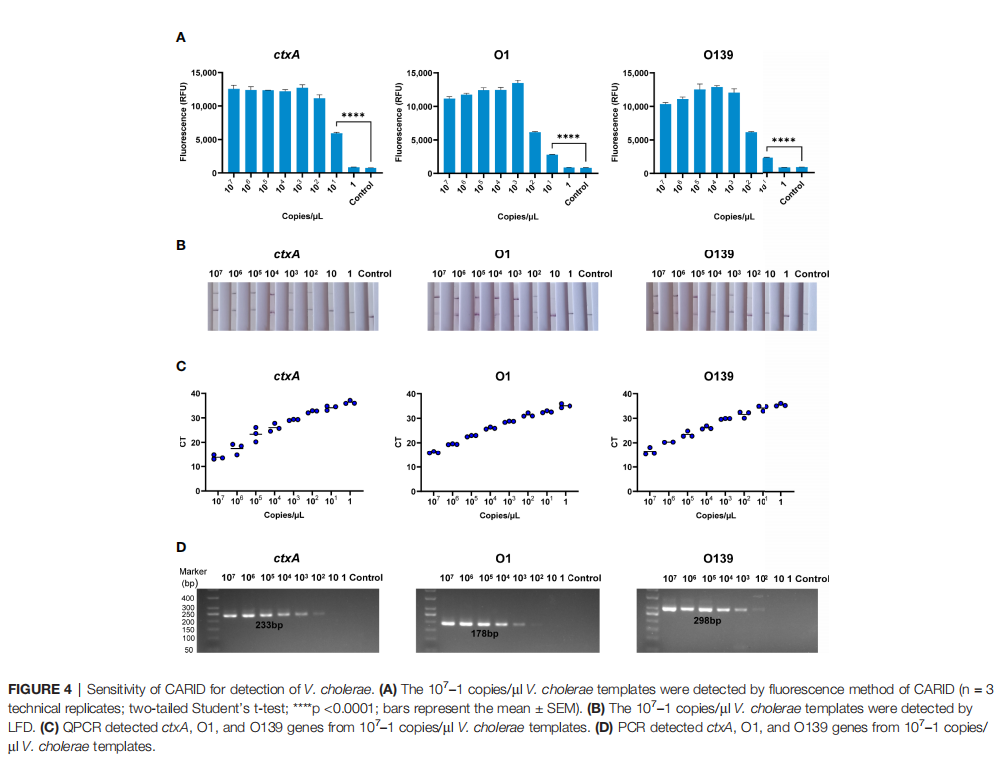
• Sensitivity:
For V. cholerae genomic DNA, the Limit of Detection (LoD) of CARID was as low as 20 copies/reaction.
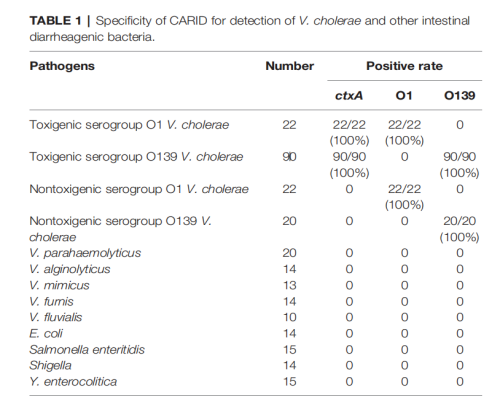
• Specificity:
The assay correctly identified all target strains with 100% accuracy. No cross-reactivity was observed when tested against diarrheal pathogens such as Escherichia coli, Salmonella Enteritidis, Shigella, Yersinia enterocolitica, and other non-target Vibrio species.
• Simulated Sample Validation
• Water Samples: The study used simulated river water samples for testing. After sample preparation, enrichment was performed in alkaline peptone water at 37°C for 4 hours, with sampling every hour. Nucleic acid was extracted from the collected samples using a rapid boiling method. After brief enrichment, CARID could detect bacterial concentrations as low as 1 CFU/mL in the river water samples.
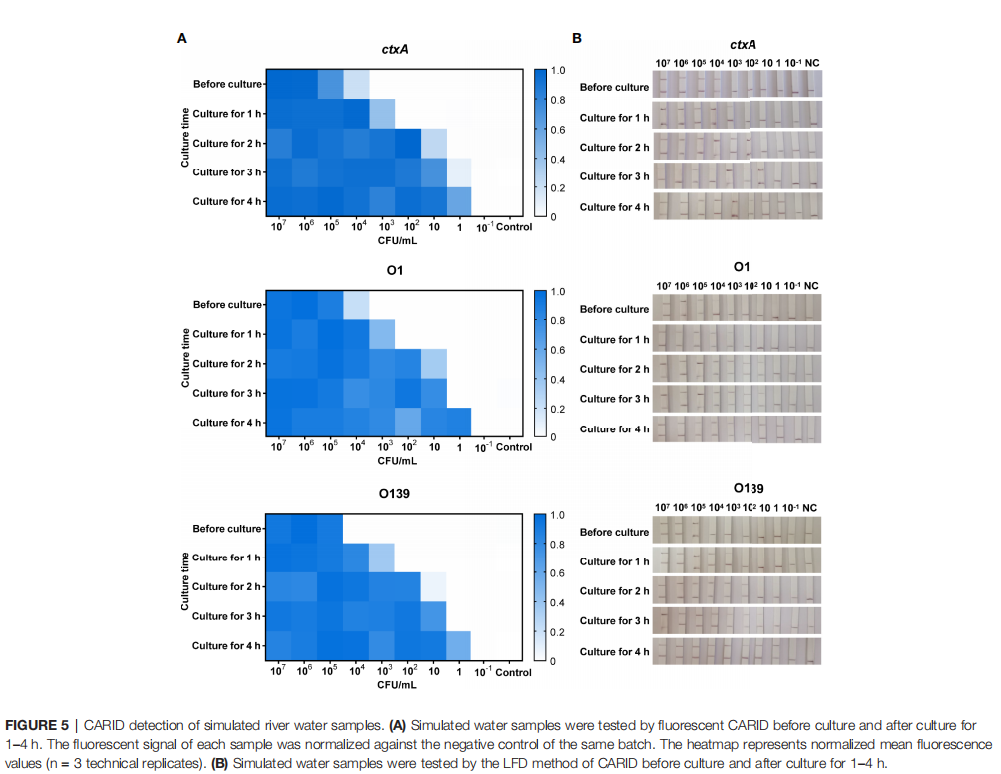
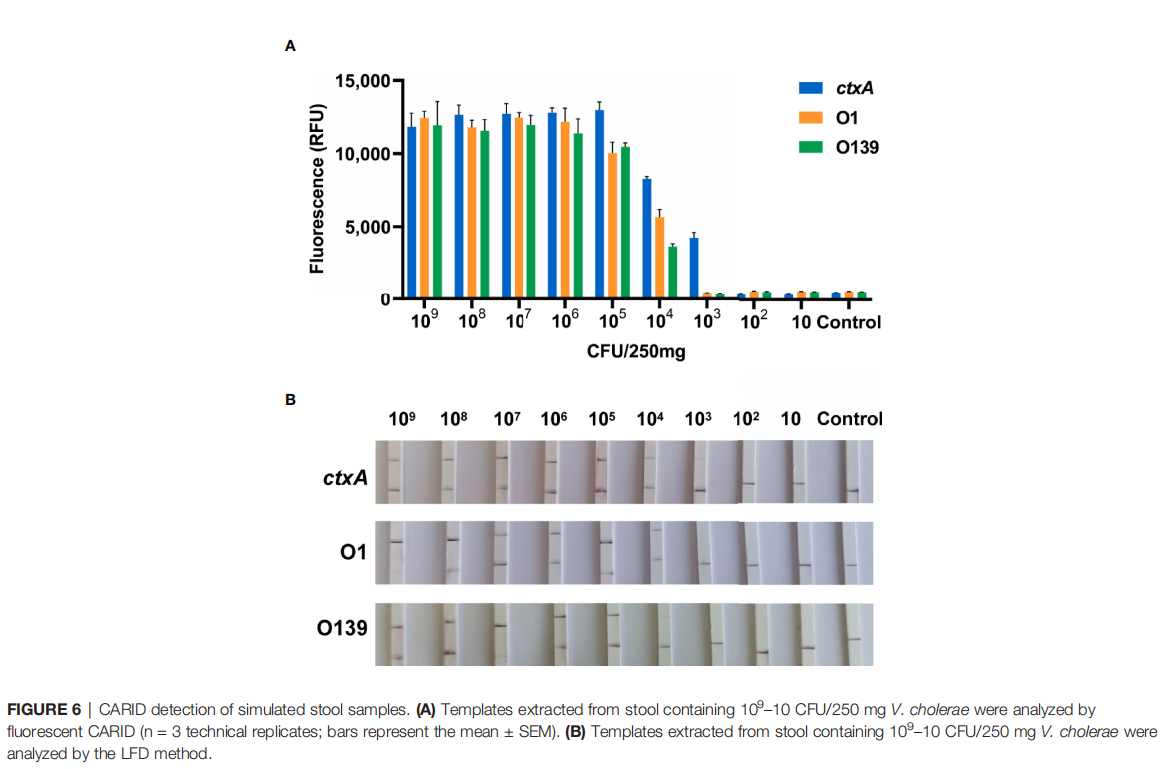
• Stool Samples: For stool samples, nucleic acid was extracted using the QIAamp® PowerFecal® Pro DNA Kit. In artificially spiked healthy human stool, the system's detection sensitivity reached 4×10³ CFU/g—significantly lower than the typical bacterial load in cholera patient stool (10⁷–10⁹ CFU/g), confirming its suitability for clinical diagnosis.
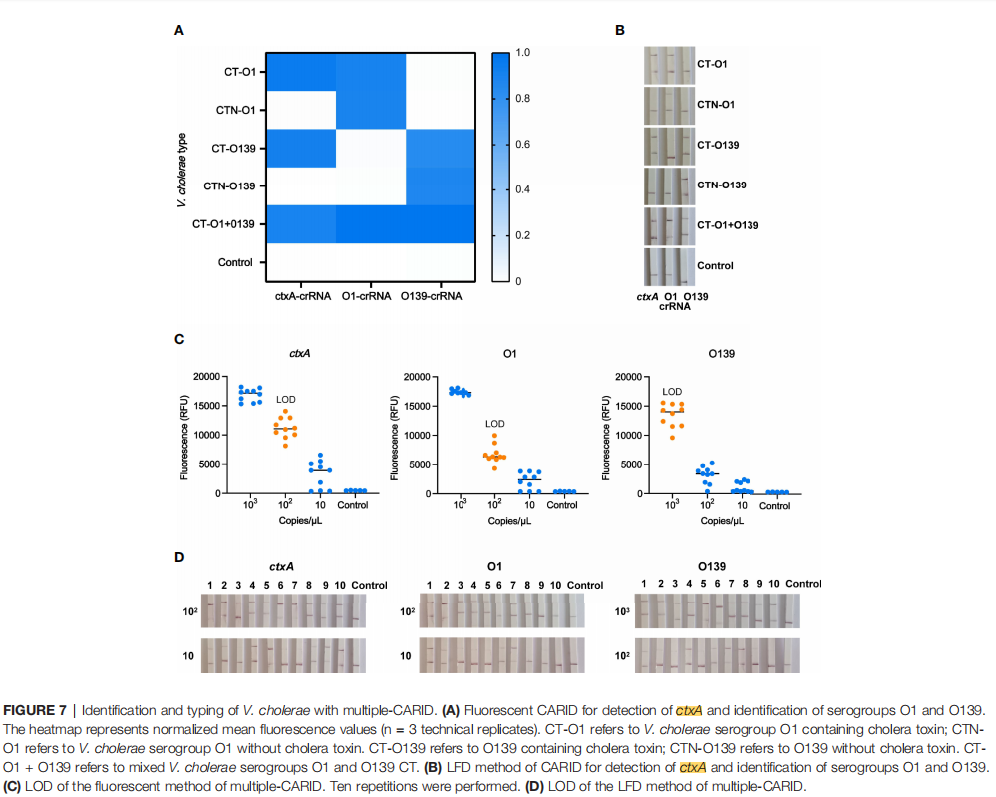
Multiplex Detection Capability
The study also developed a multiplex CARID assay capable of simultaneously detecting three key genes: the toxin gene ctxA, the O1 serogroup gene rfb-O1, and the O139 serogroup gene rfb-O139. Its Limit of Detection (LoD) was 200–2,000 copies/reaction, comparable to the singleplex assays. This multiplex approach significantly improves efficiency and saves time and costs.
Conclusion
The CARID system represents a significant advancement in the rapid, sensitive, and field-deployable detection of toxigenic Vibrio cholerae. The system can provide reliable results within one hour using low-cost equipment and visual readouts, making it a highly promising tool for cholera surveillance and outbreak response in resource-limited areas.
Note: For further information regarding the original literature, please contact qt@qt-bio.com. In addition to the CARID system mentioned in the literature, there are also more convenient and rapid nucleic acid test strip methods suitable for field detection. For details, please contact qt@qt-bio.com.
QT Biotech Co., Ltd.
Tel: +86-510-85385531
Mobile: +86-18921157475
Email: qt@qt-bio.com
Website: www.qt-bio.com
Address: No. 97, Xingye Building B, Linghu Avenue, Xiwu District, Wuxi City
Racing against time, guarding life safety

WeChat Account

Official Public Account

