News Center
News and Development
Literature Sharing: Rapid Nucleic Acid Detection of Listeria monocytogenes Based on RAA-CRISPR/Cas12a System
Release Time:
2025-05-30
CDC and public health officials in several states are investigating a multistate outbreak of Listeria infections linked to ready-to-eat foods made by Fresh & Ready Foods LLC. Foods include items like sandwiches and protein snacks.
Title: Rapid Nucleic Acid Detection of Listeria monocytogenes Based on RAA-CRISPR/Cas12a System
Authors: Yujuan Yang, Xiangxiang Kong, Jielin Yang, et al.
Journal: International Journal of Molecular Sciences (2024)
DOI: https://doi.org/10.3390/ijms25063477
Background & Significance
CDC and public health officials in several states are investigating a multistate outbreak of Listeria infections linked to ready-to-eat foods made by Fresh & Ready Foods LLC. Foods include items like sandwiches and protein snacks. (Source: CDC). Listeria monocytogenes is a highly lethal foodborne pathogen that contaminates dairy products, meats, and other foods, causing sepsis, meningitis, and a mortality rate of 27%–44%.
However, current detection methods have significant limitations:
● Traditional culture methods: Low sensitivity (prone to false negatives), time-consuming (days), and operator-dependent.
● PCR/ELISA: Requires expensive equipment (e.g., qPCR machines) and specialized training.
● LAMP, SERS, biosensors: Complex procedures, high costs, and limited accessibility.
This study integrates Recombinase-Aided Amplification (RAA) and CRISPR/Cas12a technologies to establish a rapid, highly sensitive, and cost-effective nucleic acid detection platform, offering a novel solution for food safety monitoring.
Methods
Target Selection: Designed RAA primers and crRNA targeting the conserved virulence gene hly of L. monocytogenes.
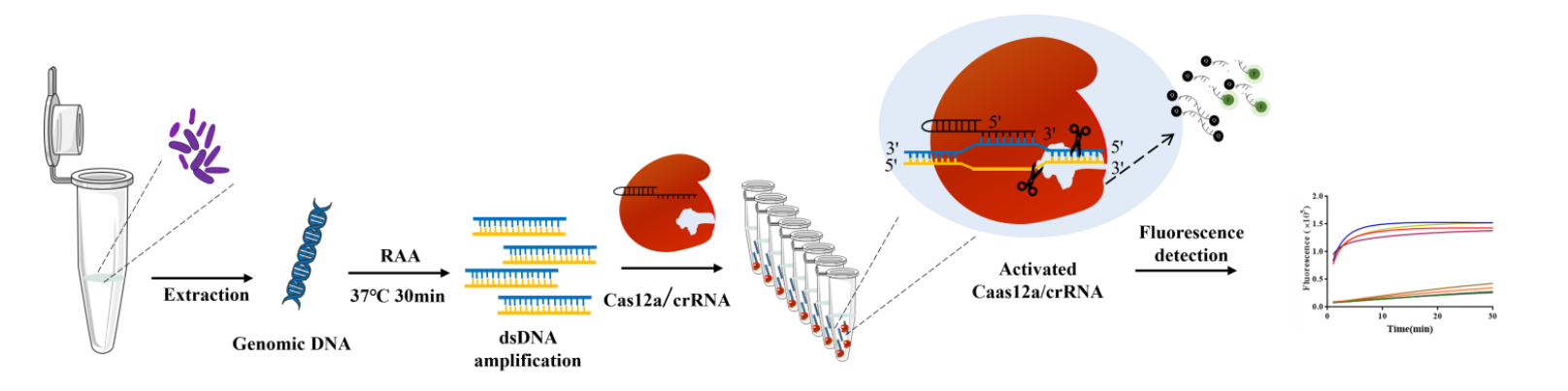
Technical Principle
● RAA Amplification (39°C, 20–30 min): Isothermal amplification of target DNA.
CRISPR/Cas12a Detection (42°C, 5 min): crRNA guides Cas12a to recognize target DNA, triggering ssDNA probe cleavage and fluorescence release (Figure 1).
Optimized Parameters
● Temperature: 42°C (higher Cas12a activity).
● Probe Ratio: Cas12a:ssDNA-FQ = 2:1 (20 μM:10 μM).
Key Results
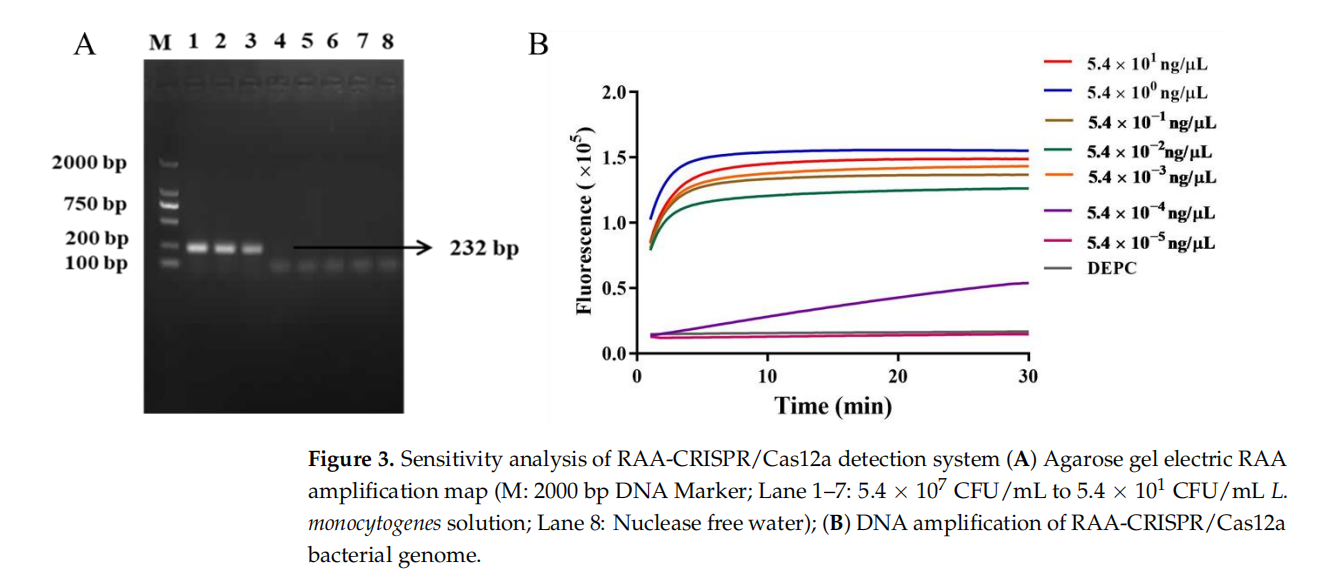
Sensitivity
● Pure bacterial culture: Detection limit of 350 CFU/mL (10× more sensitive than the national standard method).
● Genomic DNA: Detection limit of 5.4×10⁻³ ng/μL.
● Food samples (beef): Detected 2.3 CFU/25g after just 2 hours of enrichment.
Specificity
Distinguished L. monocytogenes from 5 Listeria species (e.g., L. ivanovii) and 14 common pathogens (e.g., Staphylococcus aureus, E. coli O157:H7).
Speed
Total time ≤2 hours (RAA: 30 min + CRISPR: 5 min), far faster than culture methods (3–7 days).
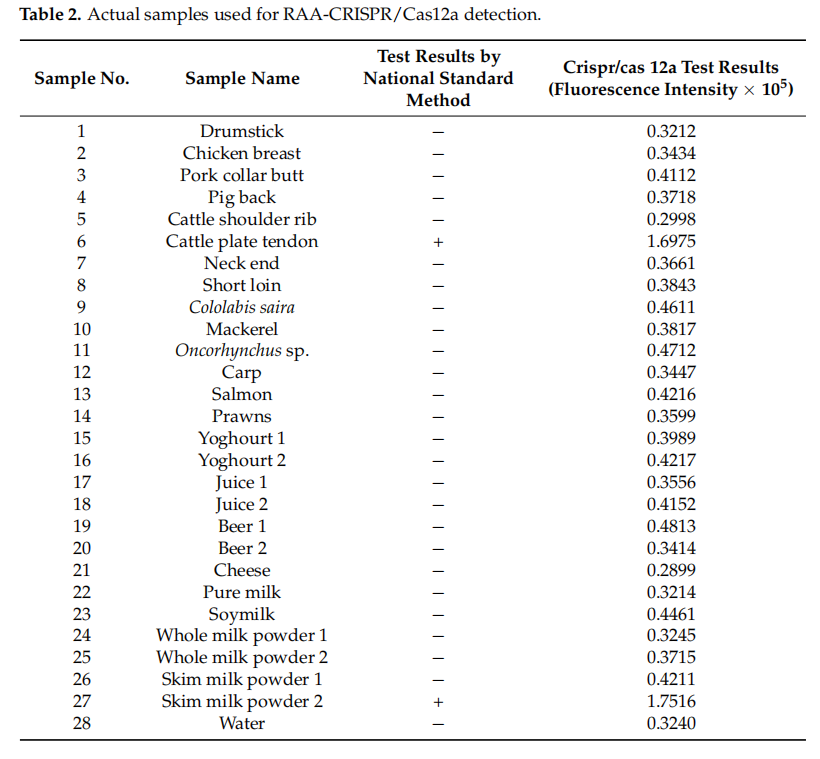
Application:
Detected 2 positives among 28 real samples (beef, milk powder, etc.), with 100% concordance with national standard methods.
Advantages
Integrated Workflow: RAA (39°C) and CRISPR (42°C) are temperature-compatible, enabling future single-tube reactions.
● Low Cost: Reaction volume of 20 μL, no need for expensive equipment (e.g., qPCR machines).
● Portability: Requires only a thermostat, suitable for field or resource-limited settings.
● Scalability: Adaptable to other pathogens (e.g., Salmonella, SARS-CoV-2) by designing new crRNAs.
Conclusion
This study provides an innovative solution for rapid, on-site screening of L. monocytogenes, particularly valuable for outbreak response and low-resource settings. The flexibility and high sensitivity of CRISPR-based diagnostics hold broad potential for food safety and clinical applications.
QT Biotech Co., Ltd.
Tel: +86-510-85385531
Mobile: +86-18921157475
Email: qt@qt-bio.com
Website: www.qt-bio.com
Address: No. 97, Xingye Building B, Linghu Avenue, Xiwu District, Wuxi City
Racing against time, guarding life safety

WeChat Account

Official Public Account

